Hi, I'm David.
I build robust, user-friendly bioinformatic tools.
I enjoy developing software that uses biological data to improve patient lives. During my PhD, I released several bioinformatic tools which focused on increasing the rate of genetic diagnoses. I found myself gripped by the problem-solving and continuous learning opportunities offered by the field of software engineering. Led by the desire to pair my bioinformatics knowledge with my interest in software engineering, I decided to move to my current role in the Biotech sector, where I have had experience in the drug discovery and diagnostics space. Always eager to dive into new languages and technologies, I explore these wherever possible in my work and through side projects; five of my favourite are displayed on this website. For a full list of my software projects and publications, check out my CV or Google scholar page.
2012
BSc Biomedical Science
2015
MSc Neuroscience
2016
Research technician
2017
PhD Bioinformatics
2022
Machine Learning Engineer
2022
Bioinformatics Software Engineer
2023
Senior Bioinformatics Software Engineer

Portfolio website
Through this side project, I took my first steps into the world of web development. My website is built using Django/Bootstrap 5, deployed with Heroku and showcases the five projects I'm most fond of.
python
HTML
CSS
Django
ggtranscript
With the increasing popularity of long-read sequencing technology came an increasing demand for tools that visualise and explore full-length transcripts. ggtranscript fills this gap by enabling users to easily create transcript annotation plots with just a few lines of code. I designed ggtranscript with the user experience in mind, extending the popular ggplot2 R package and inheriting it's functionality and familiarity.
R
Data visualisation
autogroceries
As an unadventurous, by-the-recipe cook, I had accumulated a set of ~25 go-to meals that were chosen from each week. Over time, this selection became repetitive, which motivated the development of autogroceries. autogroceries uses Selenium to automate the ordering of ingredients from the Sainsbury's website.
python
HTML
Selenium
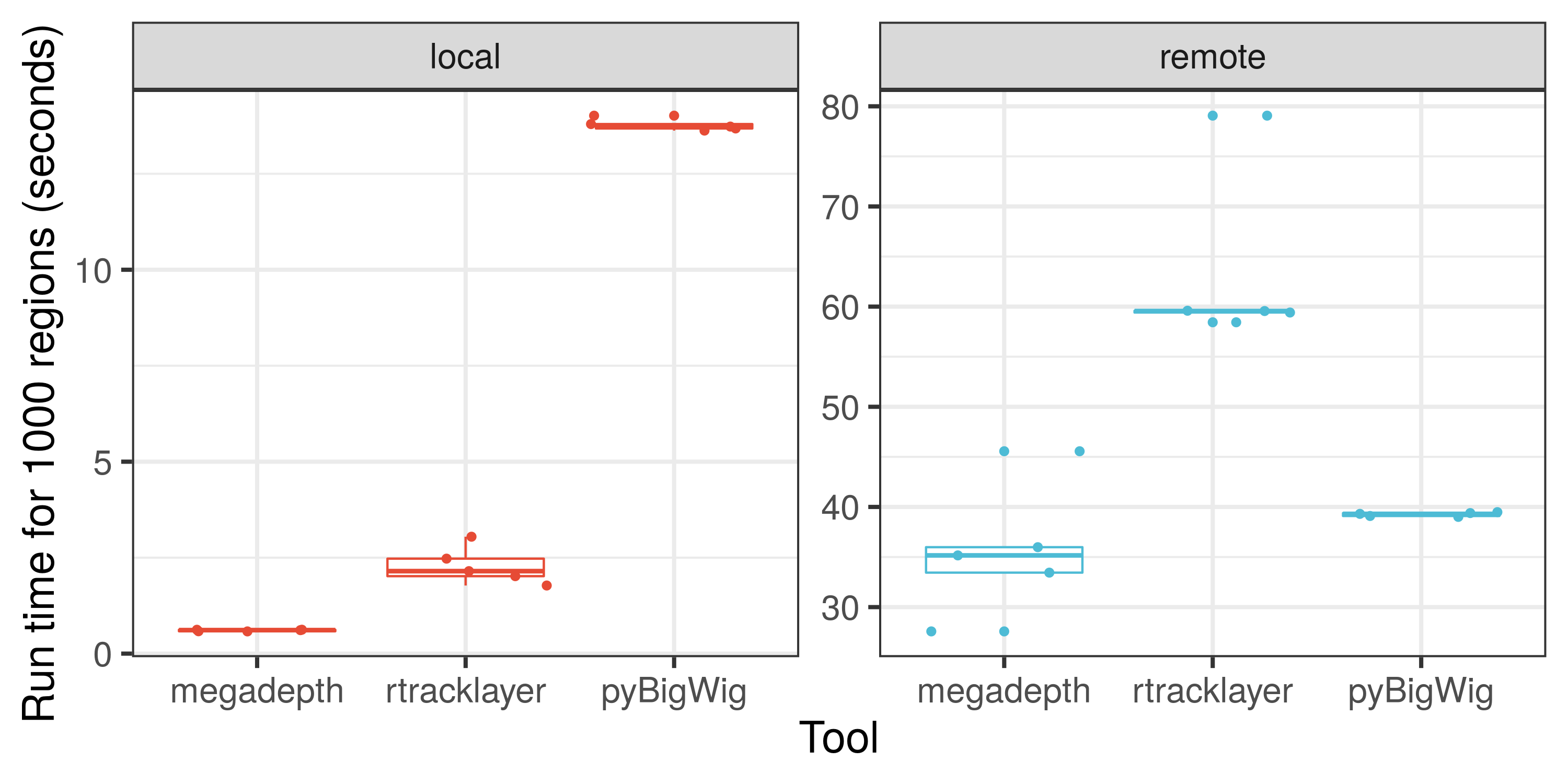
megadepth
Many bioinformatic analyses rely on quantifying coverage from BigWig/BAM files. megadepth is a tool that does this faster whilst using less memory than it's competitors. I developed the R interface for megadepth together with Leonardo Collado-Torres, which wraps the original tool created by Chris Wilks.
R